Creating a PCR Primer Table in LaTeX from a .csv File
The following example generates a nice readable LaTeX table of PCR primer sequences from a .csv file using a self defined command and csvsimple. Using this approach allows for easy storage and management of sequences (or any other data in principle) in the .csv file, without having to include and format any actual sequence data in the .tex files of the LaTeX document.
1) Getting the raw Data ready
Use a text editor or a spreadsheet editor to create a .csv file containing the gene name, forward and reverse sequence. Below is a example from my bachelor’s thesis.
The column names in the header will later be used as references to the columns.
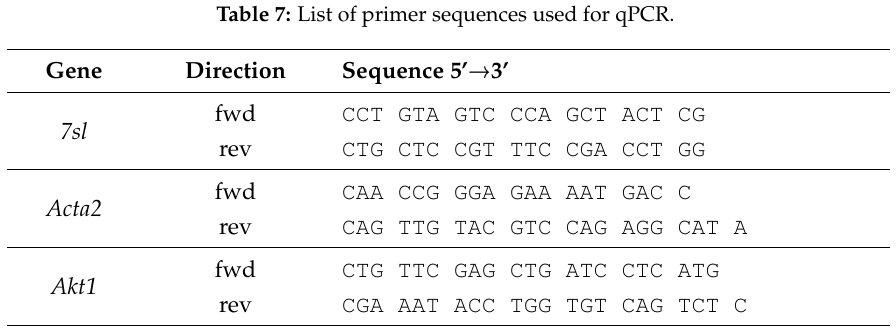
gene,seqfwd,seqrev
7sl,CCTGTAGTCCCAGCTACTCG,CTGCTCCGTTTCCGACCTGG
Acta2,CAACCGGGAGAAAATGACC,CAGTTGTACGTCCAGAGGCATA
Akt1,CTGTTCGAGCTGATCCTCATG,CGAAATACCTGGTGTCAGTCTC
2) Defining the custom Formatting Command
Inside the preamble of a latex document define a function \formatDNA{} like in the example below. When used in the document body, the function takes the provided string and inserts a space after every third character. (e.g. \formatDNA{CTGTTCGAGCT} -> CTG TTC GAG CT)
\documentclass[english,12pt,twoside]{article}
.
.
.
\makeatletter%
\newcounter{@count}%
\newcommand{\formatDNA}[1]{%
\setcounter{@count}{0}%
\@tfor\base:=#1\do{%
\base%
\stepcounter{@count}%
\ifnum\value{@count}=3
\space%
\setcounter{@count}{0}%
\fi%
}%
}%
\makeatother%
.
.
.
\begin{document}
.
.
.
3) Including the Table in the Document
In the scope of the \csvreader[]{file-to-use.csv} command, a template is defined. The command then itreates over every row of the provided file, pastes the rows data into the template and returns the resulting text.
Inside the template, the columns can be referenced using their names (e.g. \gene).
Add \usepackage[l3]{csvsimple} to your preamble.
\csvreader[head to column names]{data/qpcr_primer.csv}{}{
\\\hline \textit{\gene} & fwd \newline rev & {\ttfamily \expandafter\formatDNA\expandafter{\seqfwd} \newline \expandafter\formatDNA\expandafter{\seqrev}}
}
(\expandafter is necessary because of the order in which TeX evaluates expressions. You can read more on that here.)
When the template within the scope of the command resembles a table row, it can simply be included in a tabular (e.g. longtable) to apply additional formatting and produce a nice table in the resulting document.
This code …
\begin{longtable}{
>{\centering\let\newline\\\arraybackslash\hspace{0pt}}m{2cm}
>{\centering\let\newline\\\arraybackslash\hspace{0pt}}m{3cm}
m{9.7cm} }
\caption{List of primer sequences used for qPCR.}
\label{table:qpcr-primer}\\
\hline
\textbf{Gene} & \textbf{Direction} & \textbf{Sequence 5'\textrightarrow3'}
\csvreader[head to column names]{data/qpcr_primer.csv}{}{
\\\hline \textit{\gene} & fwd \newline rev & {\ttfamily \expandafter\formatDNA\expandafter{\seqfwd} \newline \expandafter\formatDNA\expandafter{\seqrev}}
}
\\\hline
\end{longtable}
… produces a table like the following.
Enjoy Reading This Article?
Here are some more articles you might like to read next: